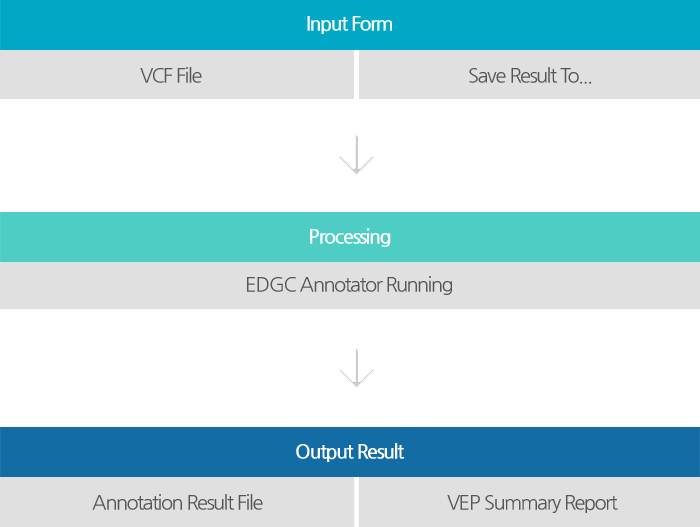
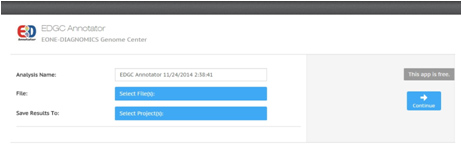
is an efficient app that annotates genetic variants of the human genome and provides information about cancer-related variants and genomic regions, allele frequency data, clinical knowledge and insights using ClinVar, OMIM, COSMIC, 1000 Genomes Project allele frequencies, dbSNP and VEP(Variant Effect Predictor) database.
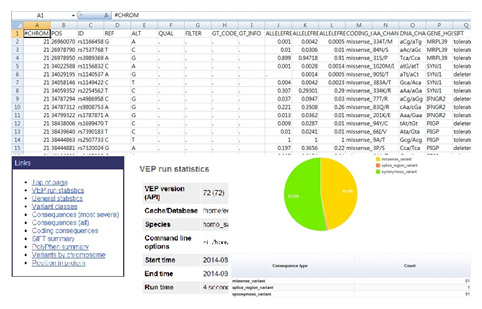
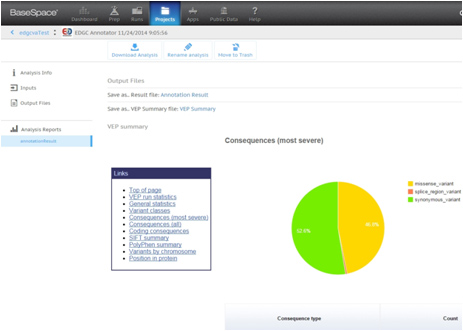
The resulting files will be provided as reports of excel and html type.